Difference between revisions of "Support:Documents:Examples:Estimate Change of Neurotransmitter"
| Line 2: | Line 2: | ||
==Overview== | ==Overview== | ||
| − | The changes of endogenous neurotransmitter by PET scanning after a stimulus has been proved with different approaches. In stead of detecting the increase or decrease of neurotransmitter, it is important to characterize the temporal change of a neurotransmitter after a stimulus. [http://www.ncbi.nlm.nih.gov/pubmed/16285909?ordinalpos=5&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_DefaultReportPanel.Pubmed_RVDocSum Morris] proposed the new technique for capturing the dynamic changes of a neurotransmitter after a stimulus and demonstrated the ability of this method to reconstruct the temporal characteistics of an enhance in neurotransmitter concentration. | + | The changes of endogenous neurotransmitter by PET scanning after a stimulus has been proved with different approaches. In stead of detecting the increase or decrease of a neurotransmitter, it is important to characterize the temporal change of a neurotransmitter after a stimulus. [http://www.ncbi.nlm.nih.gov/pubmed/16285909?ordinalpos=5&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_DefaultReportPanel.Pubmed_RVDocSum Morris] proposed the new technique (called ntPET) for capturing the dynamic changes of a neurotransmitter after a stimulus and demonstrated the ability of this method to reconstruct the temporal characteistics of an enhance in neurotransmitter concentration. |
Here we present a general model of ntPET, which can be simplified to Morris' model. | Here we present a general model of ntPET, which can be simplified to Morris' model. | ||
| Line 8: | Line 8: | ||
[[Image:Model.jpg]] | [[Image:Model.jpg]] | ||
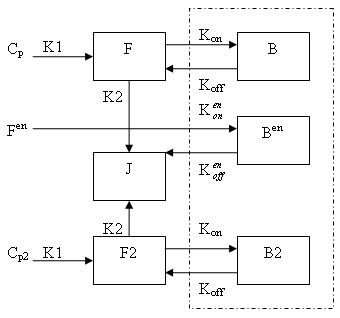
| − | Generally, there are two separate | + | Generally, there are two separate scans for ntPET: one for the rest condition (without any stimuli) and the other for the activation conditon (after a stimulus). |
Cp is the pasma concentration of tracer for the first scan (the "rest" condtion), F is free (unbound) tracer and B is bound tracer. Cp2 is the pasma concentration of tracer for the second scan (the "activation" condtion), F2 is free (unbound) tracer and B2 is bound tracer. F(en) and B(en) are free and bound neurotransmitter released by endogenous ligand. | Cp is the pasma concentration of tracer for the first scan (the "rest" condtion), F is free (unbound) tracer and B is bound tracer. Cp2 is the pasma concentration of tracer for the second scan (the "activation" condtion), F2 is free (unbound) tracer and B2 is bound tracer. F(en) and B(en) are free and bound neurotransmitter released by endogenous ligand. | ||
Revision as of 19:40, 25 February 2009
Model of Neurotransmitter PET (ntPET)
Overview
The changes of endogenous neurotransmitter by PET scanning after a stimulus has been proved with different approaches. In stead of detecting the increase or decrease of a neurotransmitter, it is important to characterize the temporal change of a neurotransmitter after a stimulus. Morris proposed the new technique (called ntPET) for capturing the dynamic changes of a neurotransmitter after a stimulus and demonstrated the ability of this method to reconstruct the temporal characteistics of an enhance in neurotransmitter concentration.
Here we present a general model of ntPET, which can be simplified to Morris' model.
Generally, there are two separate scans for ntPET: one for the rest condition (without any stimuli) and the other for the activation conditon (after a stimulus). Cp is the pasma concentration of tracer for the first scan (the "rest" condtion), F is free (unbound) tracer and B is bound tracer. Cp2 is the pasma concentration of tracer for the second scan (the "activation" condtion), F2 is free (unbound) tracer and B2 is bound tracer. F(en) and B(en) are free and bound neurotransmitter released by endogenous ligand.