Difference between revisions of "Support:Documents:Examples: Reslicing using COMKAT image tool (advanced)"
| Line 26: | Line 26: | ||
==== Create a new model ==== | ==== Create a new model ==== | ||
<pre> | <pre> | ||
| − | + | function outputVolume = fcnReSlice(GUI_handles, a_row, a_column, a_plane, flagUseCurrIvd) | |
| + | %%*****************************************************************************************%% | ||
| + | % Example: Using ImageVolumeData::sliceVolume() to reslice 3D images | ||
| + | % e.g. | ||
| + | % outputVolume = fcnReSlice(GUI_handles [, a_row, a_column, a_plane, flagUseCurrIvd]); | ||
| + | % | ||
| + | % Parameters - | ||
| + | % GUI_handles : GUI handles obtained from comkatimagetool | ||
| + | % a_row : Desired aspect ratio in row direction (default: 1.0) | ||
| + | % a_column : Desired aspect ratio in colume direction (default: 1.0) | ||
| + | % a_plane : Desired aspect ratio in plane direction (default: 1.0) | ||
| + | % flagUseCurrIvd : A flag to control the data used to reslice | ||
| + | % ( 1: Current COMKAT ImageVolumeData (default), 0: Original) | ||
| + | % outputVolume : Resliced 3D volume images | ||
| + | % | ||
| + | % | ||
| + | % Dylan Su 2012-07-30 | ||
| + | % kuan-hao.su@case.edu | ||
| + | %%*****************************************************************************************%% | ||
| − | % | + | %% Obtain the imageVolumeData object from GUI_handles |
| − | + | ivd = GUI_handles.imageVolumeData{1}; % assume PET is image 1 | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | if nargin == 1, | |
| − | + | a_row = 1; | |
| − | + | a_column = 1; | |
| + | a_plane = 1; | ||
| + | end | ||
| − | + | if nargin < 5, | |
| − | + | flagUseCurrIvd = 1; % 1: use current COMKAT image volume | |
| − | + | end | |
| − | + | if (~flagUseCurrIvd), | |
| − | + | %% Get the information from origianl data | |
| + | positionInput = get(ivd,'ImagePositionPatient'); | ||
| + | orientationInput = get(ivd,'ImageOrientationPatient'); | ||
| + | pixelSpacing = get(ivd, 'PixelSpacing'); | ||
| + | [nr, nc, np, ~] = get(ivd, 'VolumeDimension'); | ||
| + | nrows = nr; | ||
| + | ncols = nc; | ||
| + | nplanes = np; | ||
| + | |||
| + | else | ||
| + | %% Get the information from current COMKAT imageVolumeData | ||
| + | idxSA = 1; % get infor from short axis (SA) view | ||
| + | |||
| + | positionInput = GUI_handles.view{idxSA}.position{1}; | ||
| + | orientationInput = GUI_handles.view{idxSA}.orientation{1}; | ||
| + | pixelSpacing = repmat( GUI_handles.view{idxSA}.pixelSpacing(1),[1,3]); | ||
| + | nrows = GUI_handles.rows; | ||
| + | ncols = GUI_handles.columns; | ||
| + | |||
| + | [~, ~, np, ~] = get(ivd, 'VolumeDimension'); | ||
| + | |||
| + | pixelSpacingOrg = get(ivd, 'PixelSpacing'); | ||
| + | |||
| + | nplanes = ceil(np * pixelSpacingOrg(3) / pixelSpacing(3)); | ||
| + | end | ||
| − | + | %% Calculate the desired dimension | |
| − | + | dnrows = ceil(nrows * a_row); | |
| + | dncols = ceil(ncols * a_column); | ||
| + | dnplanes = ceil(nplanes * a_plane); | ||
| + | |||
| + | % calcualte new pixelSpacing by preserving the original FOV size | ||
| + | pixelSpacing = pixelSpacing ./ [dnrows/nrows, dncols/ncols, dnplanes/nplanes]; | ||
| + | |||
| + | %% Set the scale and offset the same as the original volume | ||
| + | % use same scale and offset for subvolume as original volume | ||
| + | % 0 = scaledPixel = rawPixel * s + o --> rawPixel = -o/s; | ||
| + | s = get(ivd, 'VolumeFrameBufferScaleFactor'); | ||
| + | o = get(ivd, 'VolumeFrameBufferRescaleIntercept'); | ||
| + | |||
| + | rawBackgroundPixelValue = -o/s; | ||
| + | |||
| + | %% Determine location of first pixel in output volume | ||
| + | posSA = positionInput; | ||
| + | |||
| + | % xyz of the center of first pixel | ||
| + | if (~flagUseCurrIvd), | ||
| + | % for subject's data, 'posSA' is the center of the first pixel | ||
| + | position = posSA; | ||
| + | else | ||
| + | % for imageVolumeData, 'posSA' is the center of the folume | ||
| + | position = posSA - orientationInput(:,3) * (dnplanes * pixelSpacing(3)) / 2; | ||
| + | end | ||
| + | |||
| + | planePosStep = orientationInput(:,3) * pixelSpacing(3); % calculate the step of new plane position | ||
| + | |||
| + | %% Build output volume plane-by-plane | ||
| + | outputVolume = zeros(dnrows, dncols, dnplanes); % initialize the output images | ||
| + | |||
| + | for idxP = 1 : dnplanes, | ||
| + | planePos = position + (idxP - 1) * planePosStep; % position of plane to be interpolated | ||
| + | |||
| + | % determine indicies in original volume corresponding to xyz physical (mm) location of plane | ||
| + | [u, v, w] = coordinateGen(ivd, ... | ||
| + | dncols, dnrows, pixelSpacing, planePos, orientationInput); | ||
| + | |||
| + | % obtain a slice by interpolating from the ivd volumeFrameBuffer | ||
| + | fprintf('plane ==> %i/%i\n', idxP, dnplanes); | ||
| + | outputVolume(:, :, idxP) = sliceVolume(ivd, v , u , w, rawBackgroundPixelValue,'linear'); | ||
| + | |||
| + | end | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
</pre> | </pre> | ||
Revision as of 23:20, 30 July 2012
Estimation of Physiological Parameters Using a Physiologically Based Model
Overview
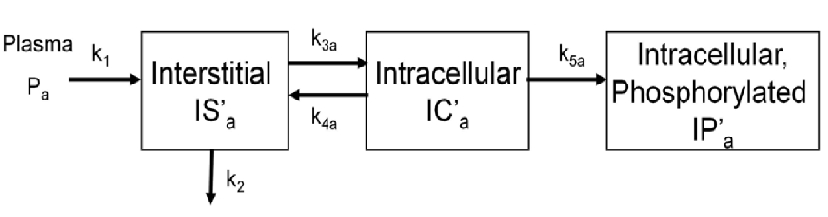
To evaluate glucose transport and phosphorylation in skeletal muscle, a physiologically based model has been proposed by our group. Herein, we show an example of implementation of the proposed model using COMKAT software. In particular, the below example shows the implementation of the proposed model for a phosphorylatable glucose analog (e.g. 18F-labeled 2-fluoro-2-deoxy-D-glucose).
The above figure shows the kinetics of [18F]2FDG in skeletal muscle. The model has three tissue compartments with five rate constants.
The first compartment is the interstitial concentration of [18F]2FDG .
The second compartment is the intracellular concentration of [18F]2FDG .
The third compartment is the intracellular concentration of [18F]2FDG-6-P .
Rate constants k1 and k2 describe the passive exchange of the glucose analog between plasma and interstitial space.
Rate constants k3a and k4a describe the inward and backward transport of the glucose analog via glucose transporters.
Rate constant k5a describe the phosphorylation of the intracellular glucose analog catalyzed by hexokinase.
COMKAT Implementation of the Physiologically Based Model
Create a new model
function outputVolume = fcnReSlice(GUI_handles, a_row, a_column, a_plane, flagUseCurrIvd)
%%*****************************************************************************************%%
% Example: Using ImageVolumeData::sliceVolume() to reslice 3D images
% e.g.
% outputVolume = fcnReSlice(GUI_handles [, a_row, a_column, a_plane, flagUseCurrIvd]);
%
% Parameters -
% GUI_handles : GUI handles obtained from comkatimagetool
% a_row : Desired aspect ratio in row direction (default: 1.0)
% a_column : Desired aspect ratio in colume direction (default: 1.0)
% a_plane : Desired aspect ratio in plane direction (default: 1.0)
% flagUseCurrIvd : A flag to control the data used to reslice
% ( 1: Current COMKAT ImageVolumeData (default), 0: Original)
% outputVolume : Resliced 3D volume images
%
%
% Dylan Su 2012-07-30
% kuan-hao.su@case.edu
%%*****************************************************************************************%%
%% Obtain the imageVolumeData object from GUI_handles
ivd = GUI_handles.imageVolumeData{1}; % assume PET is image 1
if nargin == 1,
a_row = 1;
a_column = 1;
a_plane = 1;
end
if nargin < 5,
flagUseCurrIvd = 1; % 1: use current COMKAT image volume
end
if (~flagUseCurrIvd),
%% Get the information from origianl data
positionInput = get(ivd,'ImagePositionPatient');
orientationInput = get(ivd,'ImageOrientationPatient');
pixelSpacing = get(ivd, 'PixelSpacing');
[nr, nc, np, ~] = get(ivd, 'VolumeDimension');
nrows = nr;
ncols = nc;
nplanes = np;
else
%% Get the information from current COMKAT imageVolumeData
idxSA = 1; % get infor from short axis (SA) view
positionInput = GUI_handles.view{idxSA}.position{1};
orientationInput = GUI_handles.view{idxSA}.orientation{1};
pixelSpacing = repmat( GUI_handles.view{idxSA}.pixelSpacing(1),[1,3]);
nrows = GUI_handles.rows;
ncols = GUI_handles.columns;
[~, ~, np, ~] = get(ivd, 'VolumeDimension');
pixelSpacingOrg = get(ivd, 'PixelSpacing');
nplanes = ceil(np * pixelSpacingOrg(3) / pixelSpacing(3));
end
%% Calculate the desired dimension
dnrows = ceil(nrows * a_row);
dncols = ceil(ncols * a_column);
dnplanes = ceil(nplanes * a_plane);
% calcualte new pixelSpacing by preserving the original FOV size
pixelSpacing = pixelSpacing ./ [dnrows/nrows, dncols/ncols, dnplanes/nplanes];
%% Set the scale and offset the same as the original volume
% use same scale and offset for subvolume as original volume
% 0 = scaledPixel = rawPixel * s + o --> rawPixel = -o/s;
s = get(ivd, 'VolumeFrameBufferScaleFactor');
o = get(ivd, 'VolumeFrameBufferRescaleIntercept');
rawBackgroundPixelValue = -o/s;
%% Determine location of first pixel in output volume
posSA = positionInput;
% xyz of the center of first pixel
if (~flagUseCurrIvd),
% for subject's data, 'posSA' is the center of the first pixel
position = posSA;
else
% for imageVolumeData, 'posSA' is the center of the folume
position = posSA - orientationInput(:,3) * (dnplanes * pixelSpacing(3)) / 2;
end
planePosStep = orientationInput(:,3) * pixelSpacing(3); % calculate the step of new plane position
%% Build output volume plane-by-plane
outputVolume = zeros(dnrows, dncols, dnplanes); % initialize the output images
for idxP = 1 : dnplanes,
planePos = position + (idxP - 1) * planePosStep; % position of plane to be interpolated
% determine indicies in original volume corresponding to xyz physical (mm) location of plane
[u, v, w] = coordinateGen(ivd, ...
dncols, dnrows, pixelSpacing, planePos, orientationInput);
% obtain a slice by interpolating from the ivd volumeFrameBuffer
fprintf('plane ==> %i/%i\n', idxP, dnplanes);
outputVolume(:, :, idxP) = sliceVolume(ivd, v , u , w, rawBackgroundPixelValue,'linear');
end
Specify the input, output and scan time
% Specific activity (sa): if the unit of image data is the same with that of input function, the specific activity is 1.
cm=addParameter(cm,'sa',1);
% Usually, the decay time correction of image data is performed. So, dk is zero.
cm=addParameter(cm,'dk',0);
% Define scan time (minutes)
delay = 0.0;
scanduration = 120;
t=[ones(5,1)/30;ones(10,1)/12;ones(12,1)*0.5;ones(8,1);ones(21,1)*5];
scant = [[0;cumsum(t(1:(length(t)-1)))] cumsum(t)];
scanTime = [scant(:,1),scant(:,2)];
cm = set(cm, 'ScanTime', scanTime);
lambda = [-12.02 -2.57 -0.02];
a = [1771.5 94.55 14.27];
cm = addParameter(cm, 'pfeng', [delay a lambda]');
% Define input function
cm = addInput(cm, 'Cp', 'sa', 'dk', 'fengInputByPar','pfeng'); % Cp is the plasma input function
cm = addInput(cm, 'Ca',1,0, 'fengInputByPar', 'pfeng'); % Ca is the decay-corrected whole-blood input function. Herein, we assume that Cp=Ca.
% Define output obtained from each normalized compartment
wlistTotal={'IS',1;'IC',1;'IP',1};
xlistTotal={'Ca','Fb'};
cm=addOutput(cm,'TissueTotal',wlistTotal,xlistTotal);
Usually, the model outputs consist of compartments and weights with which they contribut to outputs (e.g. Fis*IS, Fic*IC, Fic*IP).
However, as described in detail in our published paper, tracer concentrations of all compartments (i.e. IS, IC and IP) in final model equations are normalized to total tissue space.
Accordingly, the weights of these compartment are 1.
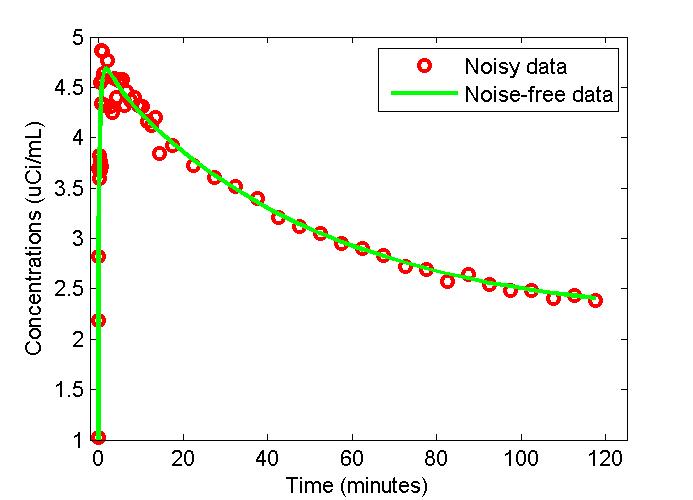
Solve the model output and Generate synthetic data
[PET,PETIndex,Output,OutputIndex]=solve(cm);
noise_level=0.05;
sd=noise_level*sqrt(PET(:,3)./(PET(:,2)-PET(:,1)));
data=sd.*randn(size(PET(:,3)))+PET(:,3);
cm=set(cm,'ExperimentalData',data);
cm = set(cm,'ExperimentalDataSD',sd);
figure;
t = 0.5*(PET(:,1)+PET(:,2));
plot(t,PET(:,3),'g-',t,data,'ro','LineWidth',2);
xlabel('Time (minutes)');
ylabel('Concentrations (uCi/mL)');
legend('Noisy data','Noise-free data');
Fit Data
% Define model parameters to be estimated
cm=addSensitivity(cm,'k1','ISg','ICg','Fis','Fb');
optsIRLS = setopt('SDModelFunction', @IRLSnoiseModel);
cm = set(cm, 'IRLSOptions', optsIRLS);
oo = optimset('TolFun', 1e-8, 'TolX', 1e-4,'Algorithm','interior-point');
cm = set(cm, 'OptimizerOptions', oo);
% Set initial conditions and bounds
% k1, ISg, ICg, Fs, Fb
pinit = [0.01 ; 4.4 ; 0.1 ;0.15 ; 0.01];
plb = [0.001; 1 ; 0.001;0.10 ; 0 ];
pub = [0.5 ; 6 ; 1 ;0.60 ; 0.04];
[pfit, qfit, modelfit, exitflag, output, lambda, grad, hessian, objfunval] = fitGen(cm, pinit, plb, pub, 'IRLS');
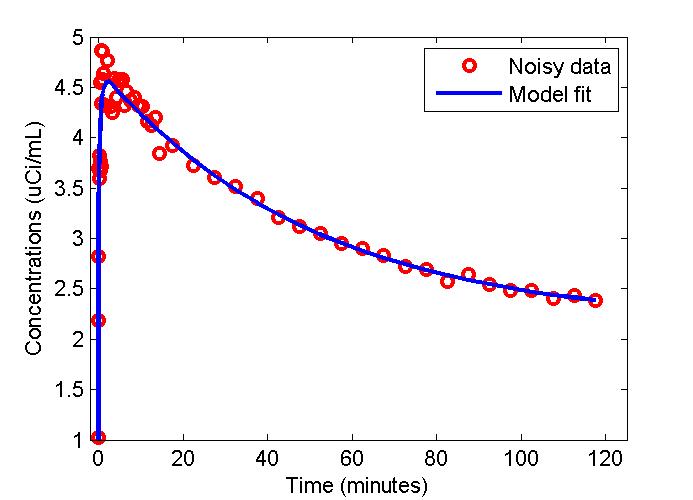
Results
figure;
t = 0.5*(PET(:,1)+PET(:,2));
plot(t,data,'ro',t,modelfit,'b-','LineWidth',2);
xlabel('Time (minutes)');
ylabel('Concentrations (uCi/mL)');
legend('Noisy data', 'Model fit');
Pg=6;KGa=14;KHa=0.17;KGg=3.5;KHg=0.13; % Units are mM
k1=pfit(1);k2=pfit(1)/pfit(4);ISg=pfit(2);ICg=pfit(3);Fis=pfit(4);Fb=pfit(5);
% Physiological parameters
VG=(k1*Pg-k1*ISg)/(ISg/(KGg+ISg)-ICg/(KGg+ICg))
VH=(k1*Pg-k1*ISg)/(ICg/(KHg+ICg))
CI=VG*ISg/(KGg+ISg)
CE=VG*ICg/(KGg+ICg)
PR=VH*ICg/(KHg+ICg)