Difference between revisions of "Support:Documents:Examples:Estimate physiological parameters using a physiologically based model"
| Line 88: | Line 88: | ||
cm=addOutput(cm,'TissueTotal',wlistTotal,xlistTotal); | cm=addOutput(cm,'TissueTotal',wlistTotal,xlistTotal); | ||
| − | % Define model parameters to be | + | % Define model parameters to be estimated |
cm=addSensitivity(cm,'k1','ISg','ICg','Fis','Fb'); | cm=addSensitivity(cm,'k1','ISg','ICg','Fis','Fb'); | ||
Revision as of 19:48, 22 August 2011
Estimation of Physiological Parameters Using a Physiologically Based Model
Overview
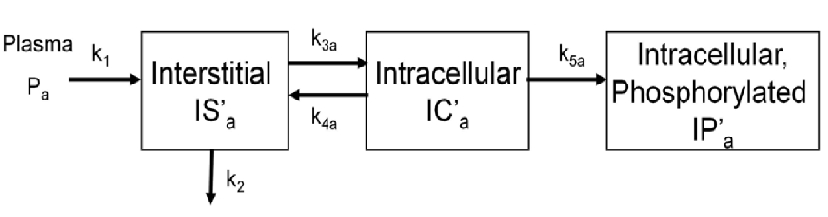
To evalute glucose trasnport and phosphorylation in skeletal muscle,a physiologically based model has been proposed by our group. The below figure shows the kinetics of a phosphorylatable glucsoe analog (e.g. 18F-labeled 2-fluoro-2-deoxy-D-glucose) in skeletal muscle. The model has three tissue compartments with five rate constants.
Rate constants k1 and k2 describe the passive exhange of the glucose analog between plasma and interstitial space.
Rate constants k3a and k4a describe the inward and backward transport of the glucose analog via glucose transporters.
Rate constant k5a describe the phosphorylation of the intracellular glucose analog catalyzed by hexokinase.
Example of Implementation of the Physiologically Based Model
Here, we show an example of implementation of the proposed model using COMKAT.
cm=compartmentModel;
% Define parameters
cm=addParameter(cm,'k1',0.1); % Diffusion of glucose analog from plasma to interstitial space
cm=addParameter(cm,'k2','k1/Fis'); % Diffusion of glucose analog from interstitial space to plasma
cm=addParameter(cm,'k3','VG/(Fis*KGa+Fis*ISg*KGa/KGg)'); % Inward transport of glucose analog
cm=addParameter(cm,'k4','VG/(Fic*KGa+Fic*ICg*KGa/KGg)'); % Backward transport of glucose analog
cm=addParameter(cm,'k5','VH/(Fic*KHa+Fic*ICg*KHa/KHg)'); % Phosphorylation of glucose analog
cm=addParameter(cm,'Fb',0.02); % fraction of total space occupied by blood
cm=addParameter(cm,'Fis',0.3); % fraction of total space occupied by interstitial space
cm=addParameter(cm,'Fic','1-Fis-Fb'); % fraction of total space occupied by intracellular space
cm=addParameter(cm,'F',1);
cm=addParameter(cm,'Pg',6); % Plasma glucose concentration (mM)
cm=addParameter(cm,'ISg',5.4); % Interstitial glucose concentration (mM)
cm=addParameter(cm,'ICg',0.2); % Intracellular glucose concentration (mM)
cm=addParameter(cm,'KGg',3.5); % Michaelis constant (mM) of glucose transporter (GLUT) for glucose
cm=addParameter(cm,'KHg',0.13); % Michaelis constant (mM) of hexokinase (HK) for glucose
cm=addParameter(cm,'KGa',14); % Michaelis constant (mM) of GLUT for glucose analog
cm=addParameter(cm,'KHa',0.17); % Michaelis constant (mM) of HK for glucose analog
cm=addParameter(cm,'VG','(k1*Pg-k1*ISg)/(ISg/(KGg+ISg)-ICg/(KGg+ICg))'); % Maximal velocity of glucose transport for glucose and its analogs
cm=addParameter(cm,'VH','(k1*Pg-k1*ISg)/(ICg/(KHg+ICg))'); % Maximal velocity of glucose phosphorylation for glucose and its analogs
% Specific activity (sa): if the unit of image data is the same with that of input function, the specific activity is 1.
cm=addParameter(cm,'sa',1);
% Usually, the decay time correction of image data is performed. So, dk is zero.
cm=addParameter(cm,'dk',0);
% Define scan time (minutes)
delay = 0.0;
scanduration = 120;
t=[ones(5,1)/30;ones(10,1)/12;ones(12,1)*0.5;ones(8,1);ones(21,1)*5];
scant = [[0;cumsum(t(1:(length(t)-1)))] cumsum(t)];
scanTime = [scant(:,1),scant(:,2)];
cm = set(cm, 'ScanTime', scanTime);
lambda = [-12.02 -2.57 -0.02];
a = [1771.5 94.55 14.27];
cm = addParameter(cm, 'pfeng', [delay a lambda]');
% Define input function
cm = addInput(cm, 'Cp', 'sa', 'dk', 'fengInputByPar','pfeng'); % Cp is the plasma input function
cm = addInput(cm, 'Ca',1,0, 'fengInputByPar', 'pfeng'); % Ca is the decay-corrected whole-blood input function. Herein, we assume that Cp=Ca.
% Define compartment
cm=addCompartment(cm,'IS'); % interstitial
cm=addCompartment(cm,'IC'); % intracellular
cm=addCompartment(cm,'IP'); % intracellular phosphorylated
cm=addCompartment(cm,'Junk');
% Define link
cm=addLink(cm,'L','Cp','IS','k1');
cm=addLink(cm,'K','IS','Junk','k2');
cm=addLink(cm,'K','IS','IC','k3');
cm=addLink(cm,'K','IC','IS','k4');
cm=addLink(cm,'K','IC','IP','k5');
% Define output obtained from each normalized compartment
wlistTotal={'IS','F';'IC','F';'IP','F'};
xlistTotal={'Ca','Fb'};
cm=addOutput(cm,'TissueTotal',wlistTotal,xlistTotal);
% Define model parameters to be estimated
cm=addSensitivity(cm,'k1','ISg','ICg','Fis','Fb');
[PET,PETIndex,Output,OutputIndex]=solve(cm);
noise_level=0.05;
sd=noise_level*sqrt(PET(:,3)./(PET(:,2)-PET(:,1)));
data=sd.*randn(size(PET(:,3)))+PET(:,3);
cm=set(cm,'ExperimentalData',data);
cm = set(cm,'ExperimentalDataSD',sd);
optsIRLS = setopt('SDModelFunction', @IRLSnoiseModel);
cm = set(cm, 'IRLSOptions', optsIRLS);
oo = optimset('TolFun', 1e-8, 'TolX', 1e-4,'Algorithm','interior-point');
cm = set(cm, 'OptimizerOptions', oo);
% Set initial conditions and bounds
% k1, ISg, ICg, Fs, Fb
pinit = [0.01 ; 4.4 ; 0.1 ;0.15 ; 0.01];
plb = [0.001; 1 ; 0.001;0.10 ; 0 ];
pub = [0.5 ; 6 ; 1 ;0.60 ; 0.04];
[pfit, qfit, modelfit, exitflag, output, lambda, grad, hessian, objfunval] = fitGen(cm, pinit, plb, pub, 'IRLS');
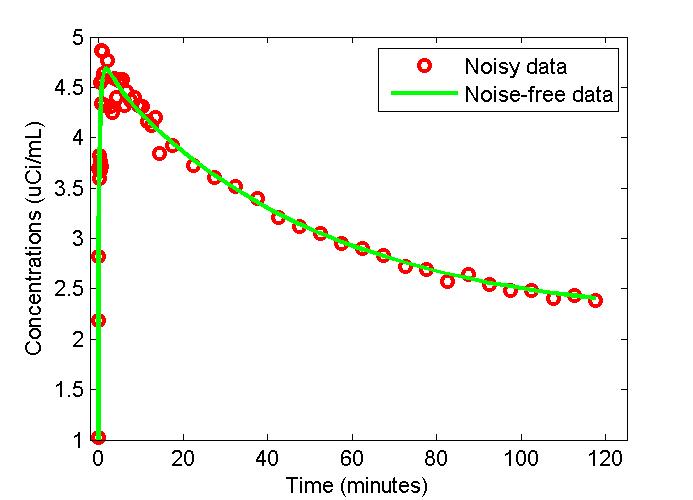
figure;
t = 0.5*(PET(:,1)+PET(:,2));
plot(t,PET(:,3),'g-',t,data,'ro','LineWidth',2);
xlabel('Time (minutes)');
ylabel('Concentrations (uCi/mL)');
legend('Noisy data','Noise-free data');
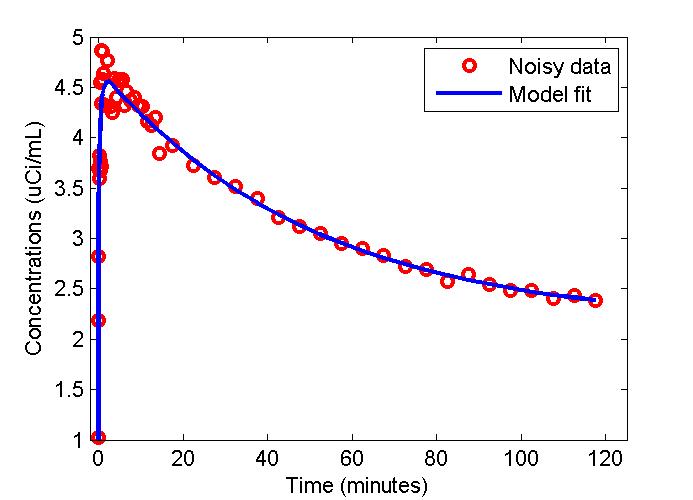
figure;
t = 0.5*(PET(:,1)+PET(:,2));
plot(t,data,'ro',t,modelfit,'b-','LineWidth',2);
xlabel('Time (minutes)');
ylabel('Concentrations (uCi/mL)');
legend('Noisy data', 'Model fit');
Pg=6;KGa=14;KHa=0.17;KGg=3.5;KHg=0.13; % Units are mM
k1=pfit(1);k2=pfit(1)/pfit(4);ISg=pfit(2);ICg=pfit(3);Fis=pfit(4);Fb=pfit(5);
% Physiological parameters
VG=(k1*ISg-k1*Pg)/(ICg/(KGg+ICg)-ISg/(KGg+ISg))
VH=(k1*Pg-k1*ISg)/(ICg/(KHg+ICg))
CI=VG*ISg/(KGg+ISg)
CE=VG*ICg/(KGg+ICg)
PR=VH*ICg/(KHg+ICg)